Life Science & Soft Condensed Matter

Read about future science
Exploring the powerhouse of lifeLife science research faces numerous challenges in the study of biological processes that occur on the atomic to cellular scale, such as large macromolecular complexes, the function of water in enzyme mechanisms and drug-substrate-product binding, and the role of biological macromolecules in membranes.
These processes are represented in diverse applications, from biofuels to cancer research.
Neutrons are ideal probes for the study of biological samples as they are very sensitive to Hydrogen-rich materials. Neutrons are even better reporters on H’s isotope, deuterium (D), and this feature is exploited by subjecting biomolecules to H/D exchange. To date, the application of neutrons to life science has been hampered by three factors:
- The low number of dedicated life science instruments
- The low neutron flux of existing sources, requiring large sample volumes and long data collection times
- Underdeveloped support for sample preparation of H/D-exchanged (or perdeuterated) molecules
With unparalleled neutron flux and sophisticated support facilities, the European Spallation Source will open the field of life sciences to neutrons and make an entirely new set of scientific experiments possible. There are several instruments that will be either dedicated to or excellent for life science studies, from single-crystal diffractometers to small-angle neutron scattering instruments.
Science Drivers
Many soft materials have commercial or medical applications for example in food technology, personal and home care products, pharmaceutical and functional materials used in a wide range of technologies. Designing and predicting the complex behaviour of these materials requires understanding the roles of individual components, for which neutron scattering techniques exploiting non-invasive deuterium labeling are an ideal tool.
Soft condensed matter encompasses a wide range of nano- and micro-structured materials based typically on polymer and surfactant systems, such as colloidal solutions, emulsions and interfacial films, including the colloidal behaviour and nanostructure of biological molecules and biomaterials. Soft matter systems contain a significant fraction of water are relatively disordered in nature, so the challenge lies in characterising their structure and dynamics on the nanometre to micrometre scales. The key techniques that address these problems are small angle neutron scattering, reflectometry, neutron spin-echo spectroscopy and quasi-elastic and vibrational neutron spectroscopy.
Solution and bulk phase structures
SANS measurements have traditionally been used to link theoretical models of fundamental colloidal interactions to the structure of novel soft materials, such as ultra-soft tunable polymer colloids[1]. Today many state-of-the-art applications investigate non-equilibrium structures and material responses to external stimuli. For example, the structures formed by colloidal suspensions under shear or flow are of significant interest for the manufacturing of personal care formulations and foods. Small angle neutron scattering can be used to collects patterns collected across a flow cell mimicking typical processing conditions to reveal coexisting structures with characteristic SANS patterns at different positions across the flow profile, typically at mm intervals[2]. Such studies require small, high intensity beams of neutrons to enable spatially resolved measurements[3] and the mapping of flow fields [4][5]. Time resolved studies of the formation and shape transformations of micelles [6][7] on the other hand allow us to probe the fundamental interactions of surfactants, and rely on manipulation of the samples using in situ equipment integrated into the neutron instrument. Many complex materials such as polymer hydrogels [8] also have hierarchical structures on several length scales from 5Å up to several micrometres and need measurements across a wide simultaneous Q-range. These applications will be addressed by the Small angle neutron scattering instruments.
Thin films at interfaces
The advanced thin film materials studied with reflectometry are becoming increasingly complex and exhibit time-dependent processes and reactions on the ms-s timescale, such as oxidation of organic films at the air-water interface by atmospheric ozone [9], or the fast swelling transition of polymer hydrogel films that can be used as a nano-scale switch in miniaturised sensors [10]. Fast low resolution studies of this kind will be one of the main strengths of the Horizontal reflectometer which will cover a broad q-range simultaneously at this time-scale. Liquid-liquid interfaces (for example oil-water[11]), are an example of significant interest in emulsion technologies, where the ESS flux will enable a significant reduction in the experimental path length, allowing experiments through bulk liquids which strongly attenuate neutrons. Minimising the beam size down to 0.3-1 mm on the Vertical reflectometer will enable studies of very small samples typical in advanced device materials, curved interfaces and scanning of local structures.
FREIA
Liquids Reflectometer
Structured and patterned surfaces
To fully understand thin 2-3D structures formed at interfaces, for example block copolymers at nano-patterned interfaces [12] or surface induced colloidal crystals [13], it is necessary to use SANS, grazing incidence SANS (GISANS) and off-specular reflectometry in combination. The ESS will be particularly well placed to further advance these applications on the Surface scattering instrument, which will access buried structures at length scales below the optical diffraction limit with chemical sensitivity and better resolution than transmission X-ray or electron microscopies. Spin-echo encoding of polarised neutrons will give access to longer length scales up to micrometres, found in many hierarchical materials.
Polymer dynamics
Many technological applications involve tailoring the dynamic properties of advanced polymer materials, for example the relationship between ion transport and polymer chain dynamics in lithium batteries[14], which can be investigated by neutron spin echo (NSE) and backscattering spectrometers covering the time window 1 picosecond - 100 nanoseconds. Neutron vibrational spectroscopy on the other hand allows observation of polymer dynamics at low frequencies without optical selection rules, for example to monitor the uptake and conformation of polyethylene oxide intercalated between graphite oxide layers used in graphene templating [15]. At ESS, such experiments will be possible with considerably less material than today, allowing straightforward characterisation of novel materials.
MAGiC
Magnetism Single-Crystal Diffractometer
Water dynamics in soft materials
Water plays an important role in the properties of soft materials and its dynamics can be probed by quasi-elastic neutron spectroscopy (QENS) on picosecond to nanosecond timescales. Deuterium labeling allows separation of the self-motions of hydrogens from the collective motions of the soft matter matrix. For example, understanding the diffusivity of water is key to determining the drug release characteristics from novel injectable drug delivery media made from thermoresponsive polymers micro-capsules[16], or the effects of soft confinement on water dynamics in polyamide engineering plastics[17], which determines their physical properties.
References
[1] B. Lonetti, M. Camargo, J. Stellbrink, C. N. Likos, E. Zaccarelli, L. Willner, P. Lindner, and D. Richter . "Ultrasoft Colloid-Polymer Mixtures: Structure and Phase Diagram", Physical Review Letters 2011, 106, 228301.
[2] Penfold, J. and I. Tucker . "Flow-Induced Effects in Mixed Surfactant Mesophases." The Journal of Physical Chemistry B 2007, 111, 9496-9503.
[3] Eberle, A. P. R.; Porcar, L. Flow-SANS and Rheo-SANS applied to soft matter. Current Opinion in Colloid and Interface Science 2012, 17, 33-43.
[4] McLeish, T. C. B.; Clarke, N.; de Luca, E.; Hutchings, L. R.; Graham, R. S.; Gough, T.; Grillo, I.; Fernyhough, C. M.; Chambon, P. Neutron flow-mapping: Multiscale modelling opens a new experimental window. Soft Matter 2009, 5, 4426-4432.
[5] Graham, R. S.; Bent, J.; Clarke, N.; Hutchings, L. R.; Richards, R. W.; Gough, T.; Hoyle, D. M.; Harlen, O. G.; Grillo, I.; Auhl, D.; McLeish, T. C. B. The long-chain dynamics in a model homopolymer blend under strong flow: small-angle neutron scattering and theory. Soft Matter 2009, 5, 2383-2389.
[6] Gummel, J.; Sztucki, M.; Narayanan, T.; Gradzielski, M. Concentration dependent pathways in spontaneous self-assembly of unilamellar vesicles. Soft Matter 2011, 7, 5731-5738.
[7] Bressel, K.; Muthig, M.; Prevost, S.; Grillo, I.; Gradzielski, M. Mesodynamics: watching vesicle formation in situ by small-angle neutron scattering. Colloid and Polymer Science 2010, 288, 827-840.
[8] Waters, D. J.; Engberg, K.; Parke-Houben, R.; Ta, C. N.; Jackson, A. J.; Toney, M. F.; Frank, C. W. Structure and Mechanism of Strength Enhancement in Interpenetrating Polymer Network Hydrogels. Macromolecules 2011, 44, 5776-5787.
[9] Thompson, K. C., A. R. Rennie, et al. "Reaction of a Phospholipid Monolayer with Gas-Phase Ozone at the Air-Water Interface: Measurement of Surface Excess and Surface Pressure in Real Time." Langmuir 2010, 26, 17295-17303.
[10] W. Wang, G. Kaune, J. Perlich, C. M. Papadakis, A. M. Bivigou Koumba, A. Laschewsky, K. Schlage, R. Röhlsberger, S. V. Roth, R. Cubitt, and P. Müller-Buschbaum, "Swelling and switching kinetics of gold coated end-capped poly(N-isopropylacrylamide) thin films", Macromolecules 2010, 43, 2444–2452.
[11] Zarbakhsh, A., J. R. P. Webster, et al. . "Structural Studies of Surfactants at the Oil-Water Interface by Neutron Reflectometery." Langmuir 2009, 25, 3953-3956.
[12] Zhang, X.; Berry, B. C.; Yager, K. G.; Kim, S.; Jones, R. L.; Satija, S.; Pickel, D. L.; Douglas, J. F.; Karim, A. Surface Morphology Diagram for Cylinder-Forming Block Copolymer Thin Films. Acs Nano 2008, 2, 2331-2341.
[13] Hellsing, M. S.; Kapaklis, V.; Rennie, A. R.; Hughes, A. V.; Porcar, L. Crystalline order of polymer nanoparticles over large areas at solid/liquid interfaces. Applied Physics Letters 2012, 100, 221601.
[14] Hua-Gen, P., T. Madhusudan, et al. (2012). Inelastic Neutron Scattering on Polymer Electrolytes for Lithium-Ion Batteries. Polymers for Energy Storage and Delivery: Polyelectrolytes for Batteries and Fuel Cells, American Chemical Society. 1096: 67-90.
[15] Barroso-Bujans, F., F. Fernandez-Alonso, et al. . "Macromolecular Structure and Vibrational Dynamics of Confined Poly(ethylene oxide): From Subnanometer 2D-Intercalation into Graphite Oxide to Surface Adsorption onto Graphene Sheets." ACS Macro Letters 2012, 1, 550-554.
[16] Paradossi et al, "Biodegradable dextran based microgels: a study on network associated water diffusion and enzymatic degradation". Soft Matter 2012, 8, 2494-2502.
[17] Laurati, M., P. Sotta, et al. "Dynamics of Water Absorbed in Polyamides." Macromolecules 2012, 45, 1676-1687.
The study of biological systems from the atomic and molecular to organism level is crucial not only for understanding health and disease, but also for biotechnological applications.
Biological samples typically have a high hydrogen content and are easily damaged by X-rays or electrons, so neutrons have many advantages as a probe for the structure and dynamics in biological systems. The use of neutrons in this field has, however, been limited by factors such as the low source brightness that leads to a large amount of sample required, limitations of isotope labelling techniques and simply the number of instruments available. The ESS long pulse source is particularly well suited for a number of neutron techniques relevant for biological systems. In combination with modern isotope labelling techniques it will allow experiments with a far larger number of biologically interesting samples.In the following we present examples illustrating what is currently state-of-the-art, what the current challanges are and what could be done in the future.
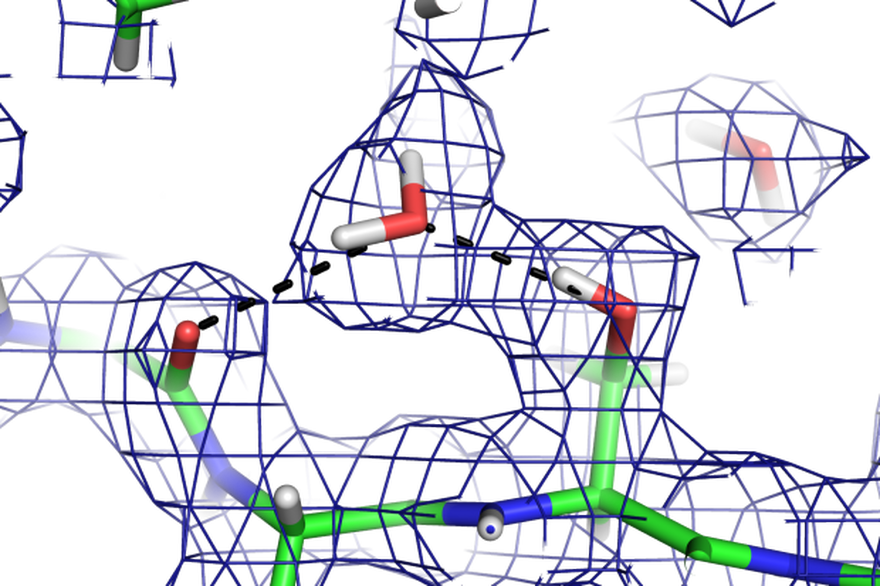
Macromolecular structures at an atomic level of detail
For structural investigations of biological macromolecules such as proteins or nucleic acids at the atomic level by crystallography the key advantage of neutrons is that they allow the hydrogen atoms to be visualised (figure). This is particularly useful for understanding phenomena such as enzyme mechanisms, protein-ligand interactions or proton transport across membranes. Understanding of enzyme mechanisms in detail is not only of fundamental chemical interest, but also essential for designing better inhibitors as pharmaceuticals and engineering industrial enzymes. A combined neutron and X-ray crystallographic study of the enzyme aldose reductase elucidated the proton transfer step in the reaction mechanism involving a deprotonated tyrosine [1]. The high brilliance of the ESS will allow similar investigations of more challenging systems such as proton pumping membrane proteins [2,3,4]. Increasing the throughput of neutron macromolecular crystallography will also enable more systematic studies of protein-ligand complexes in drug discovery. A Macromolecular Diffractometer dedicated to biological crystallography will address these challenges.
NMX
Macromolecular Diffractometer
Solution structures of macromolecular complexes
The biological function of most macromolecules involves the formation of large, multi-component complexes. Due to their inherent flexibility and transience they can often not be crystallised for structure determination. Small angle neutron scattering (SANS) can take advantage of contrast variation by changing the isotopic composition of both the solvent and the individual macromolecular components to elucidate these structures at intermediate resolution. The complex formed between myosin-binding protein C and filamentous actin was studied with SANS and contrast variation, showing how their interactions regulate contraction in the in heart muscle. [5] The high flux of the ESS will make such studies routine with smaller quantities of the protein material that is often difficult to produce for the biologically most interesting and relevant systems. The high flux could also be used to perform time-resolved studies with macromolecular complexes. A Broad-Band Small Sample SANS instrument is well adapted for such experiments.
LoKI
Broadband SANS
Biological membrane assemblies
Read about future science
Exploring the powerhouse of lifeBiological membranes are the natural environment of membrane proteins, as well as the interface between the cells and their environment. Biological membrane structures can be studied by neutron reflectometry and SANS, where deuterium labeling allows unique information about the structure across the membrane to be obtained. Observing the scattering length density profile normal the membrane surface gives information about the internal structure, such as the location [6], dimensions [7] and orientation [8] of proteins under physiologically relevant non-crystalline conditions. Time-of-flight neutron reflectometry and SANS can be used to follow molecular interactions [9], transfer [10] and reactions [11] at biological surfaces. Grazing incidence small angle scattering extends the information to the two-dimensional structure of the membrane, and the high flux of the ESS will make it possible to observe lateral structure in single biological membranes [12]. In addition to the Broad-Band Small Sample SANS instrument mentioned above, the Horizontal Reflectometer and the Surface Scattering instrument will be most useful for these investigations.
FREIA
Liquids Reflectometer
Crowding and aggregation
The cytoplasm of living cells is not a dilute solution and the various solute molecules have complicated interactions with each other and with the solvent water. The uncontrolled aggregation of macromolecules can also cause serious diseases such as Alzheimers and other neurodegenerative diseases. Early stages of protein clustering are a precursor to many such diseases, such as eye lens cataracts and amyloid plaque formation and it is thus of interest to understand the fundamental behaviour of concentrated protein solutions. SANS is ideal for determining the overall dimensions and shape of macromolecules and their complexes in concentrated solutions [13], and more transient dynamic clustering can be detected by observing the diffusion rates by Neutron Spin Echo (NSE) spectroscopy. [14] NSE experiments typically suffer from a low signal, and will considerably benefit from the ESS flux and relaxed wave-length resolution. Both a Wide-Angle and High-Resolution Spin Echo instruments, along with the Broad-Band Small Sample SANS instrument will be useful for studying macromolecular crowding and aggregation.
Dynamics of biomolecules
While techniques using elastic neutron scattering provide useful information about the average structure of biological systems, it is well-known that the dynamics also play a key role in their biological function. The motions that are of interest cover many length and time scales, from very fast dynamics on the atomic scale to the slow domain motions taking place over much longer distances. For example the changes in the vibrational dynamics of dihydrofolate reductase upon ligand binding was shown to have a significant effect on the binding free energy [15]. Other examples include water and lipid dynamics at biological interfaces [16], and large amplitude domain motions in proteins [17]. Deuterium labelling and contrast matching can be used to observe the dynamics of selective components of a complex system. Changes in the dynamical behaviour of biological macromolecules in response to external stimuli such as light in the case of the light-driven proton pump bacteriorhodopsin. These effects can be studied in specific functional states of the protein using quasielastic neutron scattering (QENS) combined with laser excitation [18]. The increase in brilliance at the ESS could be used to study ever more complex systems and changes of dynamics in a time-resolved manner to better link these changes to the biological function. To probe the range of time and length scales involved, a set of instruments including a Cold Chopper Spectrometer, a Backscattering Spectrometer as well High-Resolution and Wide-Angle Spin Echo instruments are needed.
Imaging organism-level complexity
In addition to the various neutron scattering techniques that can probe biological systems at atomic and molecular detail, neutrons can also be used to image processes at the organism level under realistic conditions. The advantages of neutrons for biological imaging are the high penetrating power, contrast between light elements and lack of radiation damage. Neutron imaging has been used for example to study the water uptake in plant roots [19], a process of obvious agricultural importance that is difficult to image with light microscopy. The ESS brilliance will make such studies more rapid, improve the time resolution and allow a more widespread use of tomographic imaging to study the three-dimensonal organisation of biological tissues and organisms in conditions not easily amenable to light microscopy. The Multi-Purpose Imaging instrument is well suited for such work.
ODIN
Multi-Purpose Imaging
References
[1] Blakeley, Matthew; Ruiz, Federico; Cachau, Raul; Hazemann, Isabelle; Meilleur, Flora; Mitschler, Andre; Ginaell, Stephan; Afonine, Pavel; Ventura, Oscar N.; Cousido-Siah, Alexandra; Haertlein, Michael; Joachimiak, Andrzej; Myles, Dean; Podjarny, Alberto (2008) "Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase", Proceedings of the National Academy of Sciences, 105, no. 6, 1844-1848
[2] Abrahams, J.P.; Leslie, A.G.W.; Lutter, R.; Walker, J.E., (1994) "Structure at 2.8 √Ö resolution of F|1|-ATPase from bovine heart mitochondria" Nature, 370, no. 6491, pp. 621-628
[3] Pedersen BP; Buch-Pedersen MJ; Morth JP; Palmgren MG; Nissen P. (2007) "Crystal structure of the plasma membrane proton pump" Nature, 450, no. 7172, pp. 1111-1114
[4] Kellosalo, Juho; Kajander, Tommi; Kogan, Kosntantin; Pokharel, Kisun; Goldman, Adrian. (2012) "The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase" Science, 337, pp. 473-476
[5] Whitten, Andrew E.; Jeffries, Cy M.; Harris, Samantha P.; Trewhella, Jill (2008) "Cardiac myosin-binding protein C decorates F-actin: Implications for cardiac function" Proceedings of the National Academy of Sciences, 105, no. 47, pp. 18360-18365
[6] Chenal, A., L. Prongidi-Fix, et al. (2009). "Deciphering Membrane Insertion of the Diphtheria Toxin T Domain by Specular Neutron Reflectometry and Solid-State NMR Spectroscopy." Journal of Molecular Biology 391(5): 872-883.
[7] Le Brun, A. P., S. A. Holt, et al. (2008). "Monitoring the assembly of antibody-binding membrane protein arrays using polarised neutron reflection." European Biophysics Journal with Biophysics Letters 37(5): 639-645.
[8] Johnson, C., G. Fragneto, et al. (2005). "Structural studies of the neural-cell-adhesion molecule by X-ray and neutron reflectivity." Biochemistry-Us 44(2): 546-554.
[9] Clifton, L. A., C. L. Johnson, et al. (2011). "Low resolution structure and dynamics of a Colicin-Receptor complex determined by neutron scattering." Journal of Biological Chemistry.
[10] Garg, S., L. Porcar, et al. (2011). "Noninvasive Neutron Scattering Measurements Reveal Slower Cholesterol Transport in Model Lipid Membranes." Biophysical Journal 101(2): 370-377.
[11] Thompson, K. C., A. R. Rennie, et al. (2010). "Reaction of a Phospholipid Monolayer with Gas-Phase Ozone at the Air-Water Interface: Measurement of Surface Excess and Surface Pressure in Real Time." Langmuir 26(22): 17295-17303.
[12] Lingwood, Daniel; Simons, Kai (2010) "Lipid rafts as a membrane-organizing principle." Science, 327, no. 5961, pp. 46-50
[13] Stradner, A.; H. Sedgwick, et al. (2004). "Equilibrium cluster formation in concentrated protein solutions and colloids." Nature 432(7016): 492-495.
[14] Falus, P.; Porcar, L.; Fratini, E.; Chen, W.-R.; Faraone, A.; Hong, K.; Baglioni, P.; Liu, Y. "Distinguishing the monomer to cluster phase transition in concentrated lysozyme solutions by studying the temperature dependence of the short-time dynamics." Journal of Physics - Condensed Matter 2012, 24, 064114
[15] Balog, Erika; Becker, Torsten; Oettl, Martin; Lechner, Ruep; Daniel, Roy; Finney, John; Smith, Jeremy C. (2004) "Direct Determination of Vibrational Density of States Change on Ligand Binding to a Protein" Physical Review Letters, 93, p. 28103
[16] Wood, K.; Pazenet, M.; Gabel, F.; Kessler, B.; Oesterhelt, D.; Tobias, D.J.; Zaccai, G.; Weik, M. (2007) "Coupling of protein and hydration-water dynamics in biological membranes" Proceedings of the National Academy of Sciences, 104, no. 46, pp. 18049-18054
[17] Pieper, Jörg; Buchsteiner, Alexandra; Dencher, Norbert A.; Lechner, Ruep E.; Hauss, Thomas (2008) "Transient Protein Softening during the Working Cycle of a Molecular Machine" Physical Review Letters, 100, p. 228103
[18] Bu, Zimei; Biehl, Ralf; Monkenbusch, Michael; Richter, Dieter; Callaway, David J.E. (2005) "Coupled protein domain motion in Taq polymerase revealed by neutron spin-echo spectroscopy" Proceedings of the National Academy of Sciences, 102, no. 49, pp. 17646-17651,
[19] Oswald, Sascha E.; Menon, Manoj; Carminati, Andrea; Vontobel, Peter; Lehmann, Eberhard; Schulin, Rainer "Quantitative Imaging of Infiltration, Root Growth, and Root Water Uptake via Neutron Radiography" Vadose Zone Journal, 7, no. 3, pp. 1035-1047