An international collaboration between the UK’s UCL School of Pharmacy, Lund Protein Production Platform at Lund University, and ESS - through it's DEMAX laboratory, have initiated bio-physical and structural studies of three non-structural proteins from the novel coronavirus, SARS CoV-2, the causative agent of COVID-19.
The Deuteration & Macromolecular Crystallisation (DEMAX) platform at ESS has been offering prioritised access to laboratory services for scientists and researchers working on COVID-19-related research projects. Under this call, a research project on viral proteins was accepted and the research team recently managed to solve, and have now started to analyse, one of these proteins, Nsp10, with the help of the BioMAX beamline at MAX IV Laboratory.
To be able to find an efficient drug, which prohibits the novel coronavirus from causing the disease COVID-19, one important aim is to understand how to block the virus from replicating its genomic material. By doing so, the virus will ‘die out’ over time as it fails to reproduce or make infectious particles. One way of obtaining this knowledge is to use X-rays to obtain an image of the structure of the proteins involved in this process.
There are 16 non-structural proteins (NSPs) in SARS CoV-2 that play important roles in viral replication and transcription. The virus has proof-reading capabilities and the proteins involved are possible drug targets.
Interfering with the virus’s ability to replicate and produce a mature, infectious virus capsid is a key focus for many international researchers. High-resolution crystallographic studies play an important role in finding either new inhibitors or, studying how existing drugs can be repurposed to block the novel coronavirus.
There are several proteins involved with proofreading of the viral RNA and its protection by RNA cap methylation, ensuring RNA replication. The research team’s studies focus on three of these: Nsp10, Nsp14, and Nsp16. A key part of this work is to establish crystallisation conditions to enable high-throughput fragment-based screening in the search for novel small molecule inhibitors that can block the viral replication cycle.
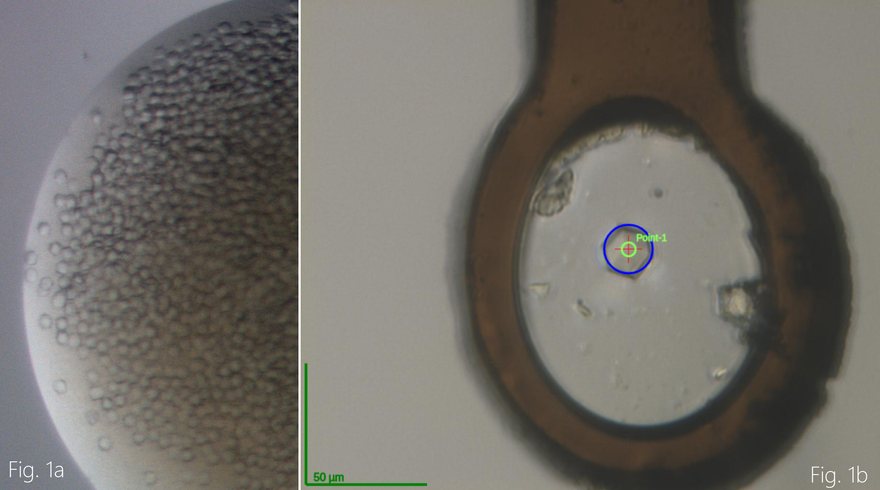
Recently, the research team had success and were able to produce crystals of one of these proteins, Nsp10 (Figure 1). Using rapid access, the data was collected remotely at the BioMAX beamline at MAX IV to approx. 2.6 Å resolution. The structure was determined and structural analysis is ongoing (Figure 2). The other work on Nsp14 and 16 also continues and the research team are working towards crystal structures of those as well.
The ultimate goal for this research project is to obtain high resolution X-ray crystal structures of these proteins alone or in complex with each other to enable the search for small molecule inhibitors that disrupt their activity. These structural studies will also be complemented with other biophysical characterisation experiments such thermal stability, microscale thermophoresis and solution scattering studies using X-rays and neutrons.
When ESS is open to neutron users, we will be able to offer researchers access to instrumentation and beamtime to pursue scientific research and investigations with the help of neutron rays. According to Dr. Zoë Fisher, group Leader for DEMAX at ESS, "Neutron protein crystallography of these viral proteins in complex with inhibitors can reveal atomic level details of hydrogen atoms and binding interactions that could be crucial for understanding - and potentially improving - on inhibitor binding. Small angle scattering of different complexes of these proteins in solution could also help understand the behaviour and shape of the molecules under physiological conditions. "
It is supported by the MAX IV rapid access call COVID-19,, and the ESS DEMAX call for prioritised projects related to COVID-19 research.